
Part of the reason for why I have such strong opinions about this matter, is that I've actually witnessed scientists waste more time messing around in fields where they were clearly out of their depth.
Most scientists also already do this implicitly by choosing to purchase instrumentation from manufacturers like Olympus, Phillips, or Siemens rather than building it themselves. This sort of delegation of course, is actually more efficient, since you can work on other tasks in parallel with the engineer (such as writing your next grant proposal or article or gasp teaching). It would also be incorrect to assume that engineering does not involve creativity and is purely bound by rigid processes- if your requirements were strange enough, something fresh would inevitably be built. If you can give an engineer general specifications for what you want and then leave him/her alone, he/she should be able to produce something that mostly fits your needs while avoiding all of the pitfalls that wouldn't have occurred to you. Indeed, there's a reason for the qualifier rapid in the term "rapid prototyping". I think that it's unrealistic of you to believe that engineers always work with "a fully dimensioned and toleranced drawing" before starting work on a project and that your would work "grind to a halt".
What I find objectionable is the inability of scientists to explicitly delegate tasks to domain specialists in their everyday work when it makes sense. I agree that science can't be bound by the rigid structures of most applied disciplines, and that the freedom to combine technologies in novel ways is a pre-requisite to novel findings.
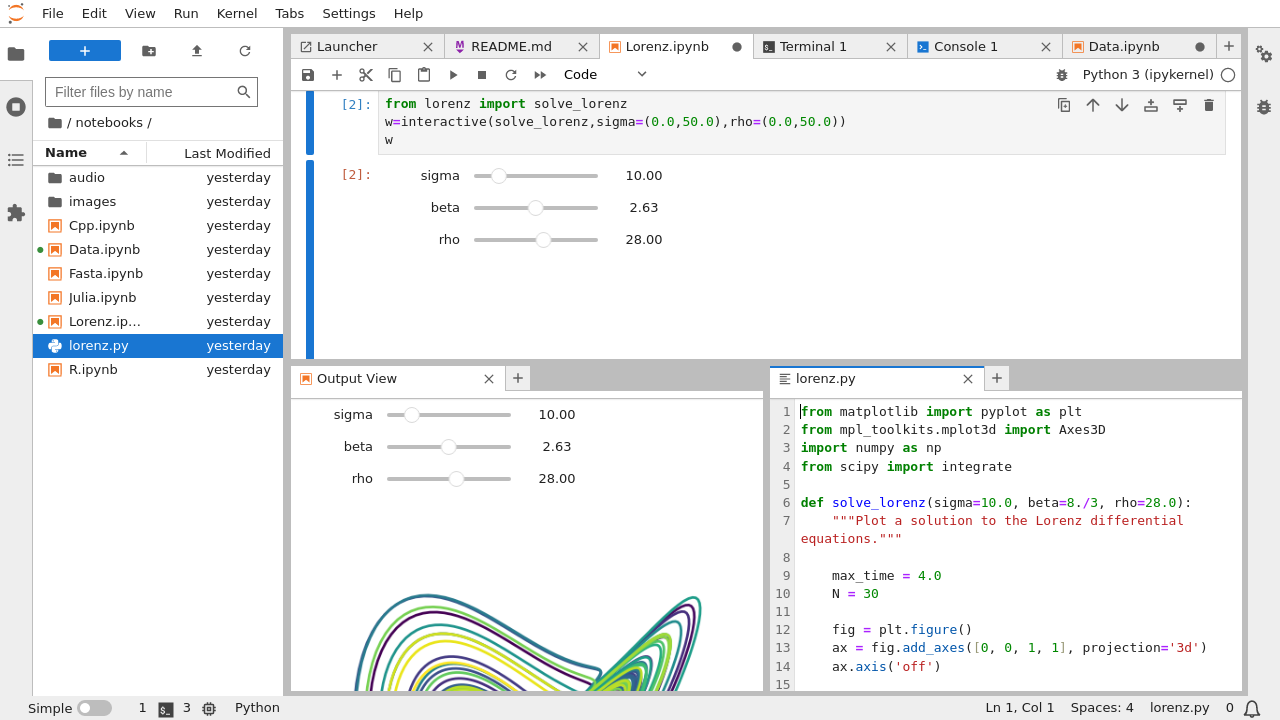
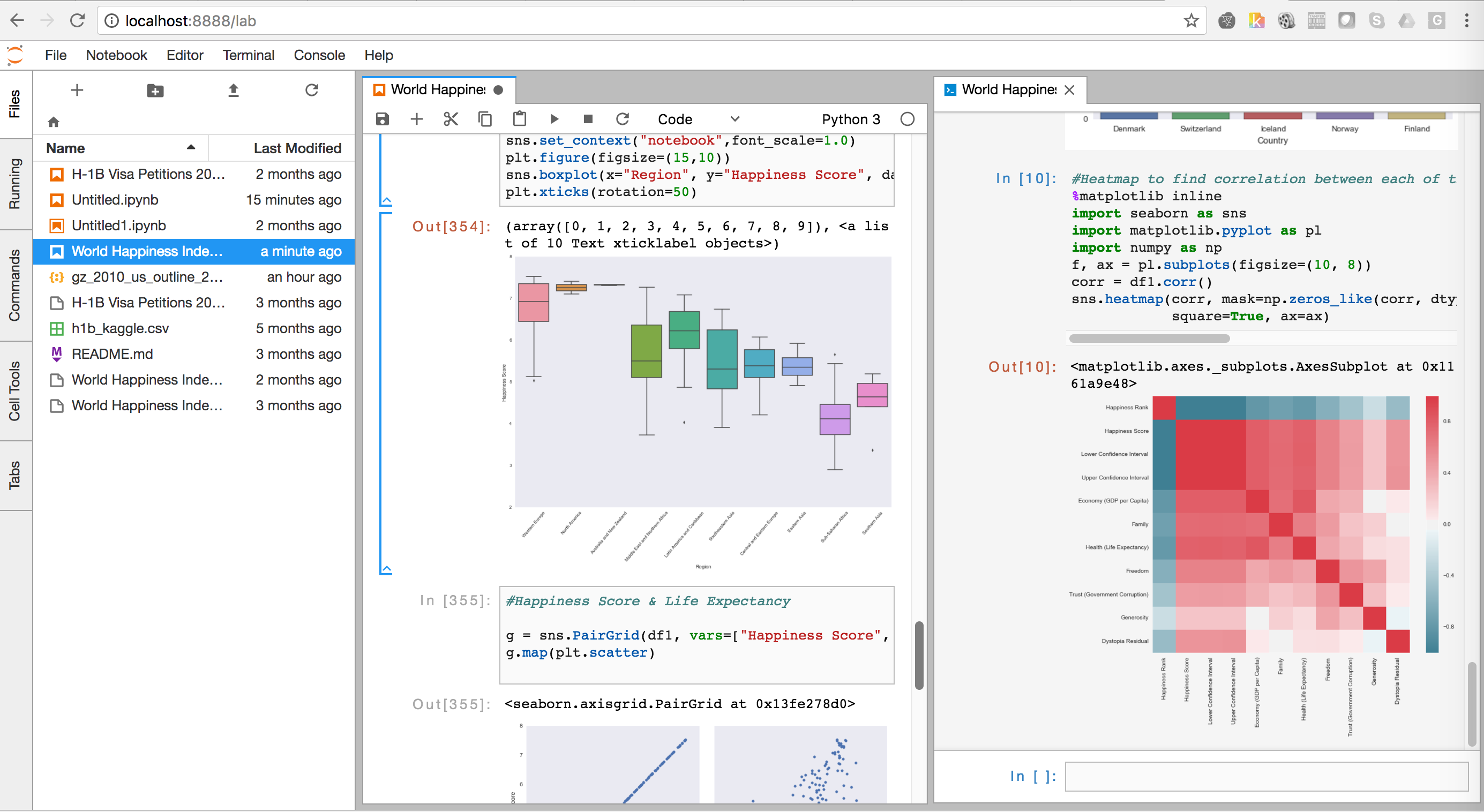
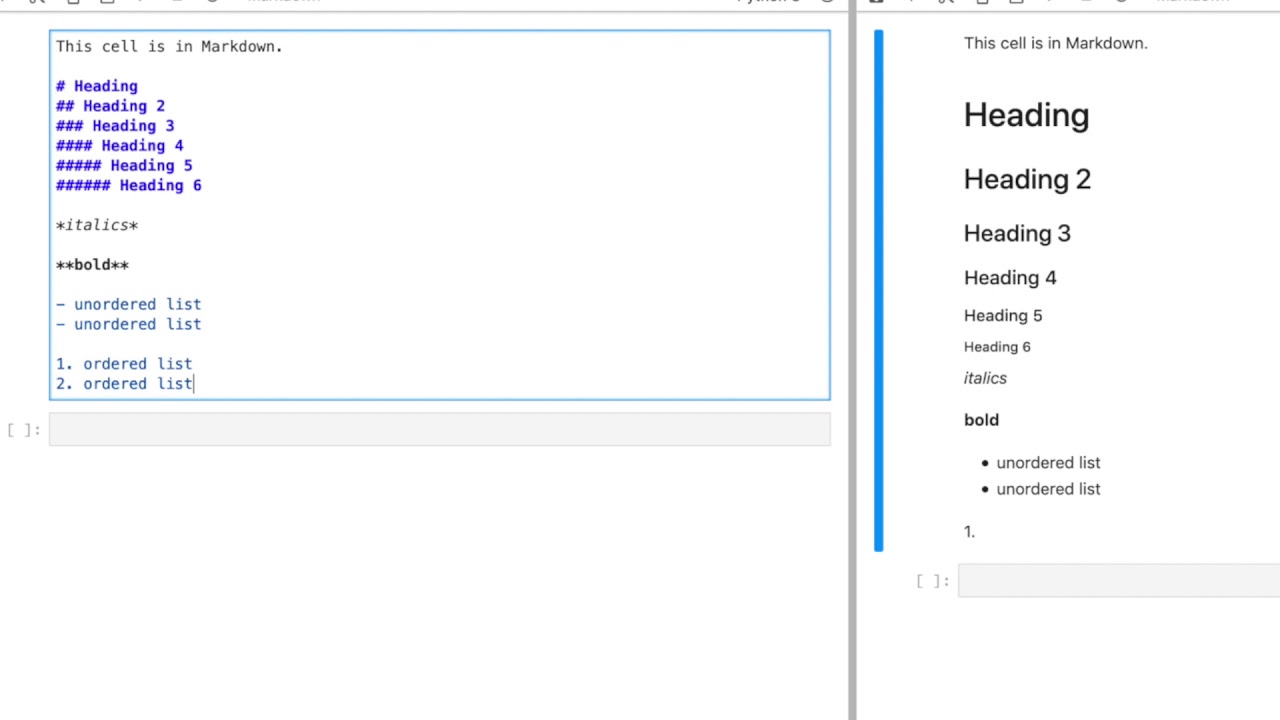
COMBINING JUPYTERLAB AND LIGHTTABLE CODE
Granted, I've seen huge notebooks that are a mess, so I understand the frustration, but it's not like we all haven't seen the single file of code with 5000 lines and 10 nested layers of conditionals at some point in our lives. When I hear "how do I import a notebook like a python module", I'm very very scared. In that regard I think it's barely an improvement over. When I see stuff around notebooks for "reproducibility", I'm a bit confused in that notebooks often don't specify any guidance on installation and dependencies, let alone things like arguments and options that a regular old script would. When I see stuff like "out of order execution is confusing", I don't disagree, but it does make me wonder how long and convoluted the notebooks these people work with are - probably a ripe candidate to refactor stuff out into python modules as functions. It is there for you to a) try things out yourself, and whatever you discover hopefully eventually make it into proper python modules b) make interesting ideas presentable and explorable for others. It's apples and oranges w.r.t and a poor substitute for something like P圜harm or VS Code or vim. It's decidedly NOT there for you to type all your code in like an editor and make a huge mess. From the document side, as a bonus, you get nicer presentation (charts, interactivity, nice and wide sortable tables, etc) than you would in a shell, which comes in handy when doing things like data exploration or mathematical simulation.
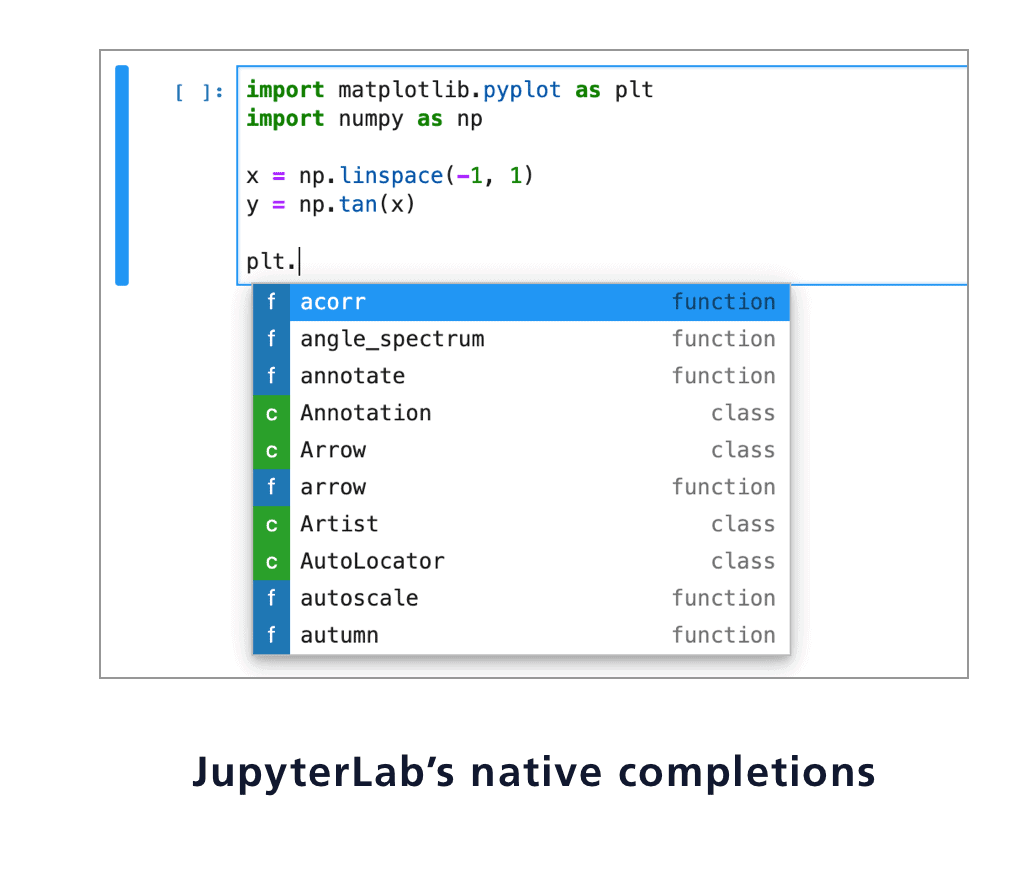
If I change this variable, how will the system react? You can just try it!ī) Just like with a REPL, you can type in and execute commands step by step, viewing the output of the previous command instead of running the whole thing at once. From the REPL side, as a bonus, you get interactivity, and the reader can pause and experiment with the examples you're giving to improve their understanding or test their hypotheses. So it's useful for a bit of both:Ī) Just like with a paper, you can present scientific or mathematical ideas with accompanying visualizations or simulations. It's a mashup between a scientific paper and a repl. The majority of the complaints I hear about notebooks I think come from a misunderstanding of what they're supposed to be.


 0 kommentar(er)
0 kommentar(er)
